Kaynağa Gözat
Add more performance feedback documentation
15 değiştirilmiş dosya ile 11417 ekleme ve 1 silme
+ 12
- 0
doc/doxygen/Makefile.am
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
BIN
doc/doxygen/chapters/data_trace.pdf
Dosya farkı çok büyük olduğundan ihmal edildi
+ 4283
- 0
doc/doxygen/chapters/distrib_data.eps
BIN
doc/doxygen/chapters/distrib_data.pdf
BIN
doc/doxygen/chapters/distrib_data.png
Dosya farkı çok büyük olduğundan ihmal edildi
+ 1613
- 0
doc/doxygen/chapters/distrib_data_histo.eps
BIN
doc/doxygen/chapters/distrib_data_histo.pdf
BIN
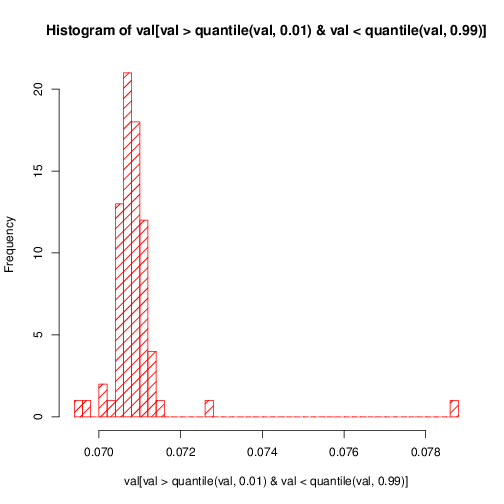
doc/doxygen/chapters/distrib_data_histo.png
+ 13
- 1
doc/doxygen/chapters/performance_feedback.doxy
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
Dosya farkı çok büyük olduğundan ihmal edildi
+ 1036
- 0
doc/doxygen/chapters/starpu_chol_model_11_type.eps
BIN
doc/doxygen/chapters/starpu_chol_model_11_type.pdf
BIN
doc/doxygen/chapters/starpu_chol_model_11_type.png

Dosya farkı çok büyük olduğundan ihmal edildi
+ 4460
- 0
doc/doxygen/chapters/starpu_non_linear_memset_regression_based_2.eps
BIN
doc/doxygen/chapters/starpu_non_linear_memset_regression_based_2.pdf
BIN
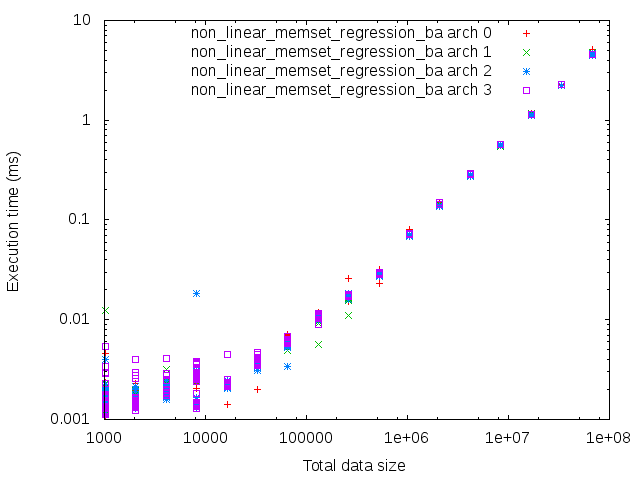
doc/doxygen/chapters/starpu_non_linear_memset_regression_based_2.png
